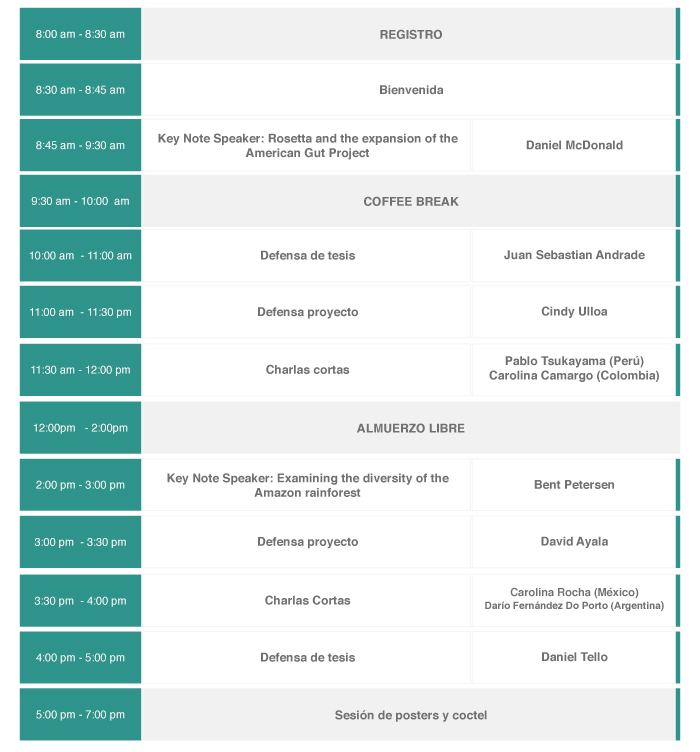
3er Foro Internacional de Biología Computacional:

El mundo está de lleno en la nueva era de la Biología Computacional. Esta rama de la Biología y de las Ciencias Computacionales tiene un rol esencial en estudios a gran escala en temas que van desde la biodiversidad y la conservación hasta la solución de problemas en salud y agricultura que pueden significar la supervivencia humana en el mundo. Cada día se producen en el mundo millones de datos en el estudio de la Biología y la Medicina y gracias a los avances computacionales es ahora factible realizar análisis que hace unos años no habrían sido posibles.
La generación de estas cantidades masivas de datos es cada vez más económica y asequible. Sin embargo, el factor limitante ahora es el personal calificado y entrenado con la capacidad de analizar e interpretar los datos. Colombia debe posicionarse en esta área en la que tiene un alto potencial de ser competitiva. En este foro se tendrán charlas de importantes expertos internacionales en el área de la metagenómica y la caracterización de comunidades y diversidad ambiental. Se complementarán con las presentaciones y defensas de tesis de los estudiantes de la Maestría en Biología Computacional así como charlas cortas de estudiantes e invitados.
*Cambios sin previo aviso
Panelistas
Daniel McDonald - University of California San Diego
Charla: Rosetta and the expansion of the American Gut Project
Position: Scientific Director, American Gut Project
Affiliation: Department of Pediatrics, School of Medicine, UC San Diego
Dr. McDonald is the Scientific Director for The Microsetta Initiative and the American Gut Project, run by the School of Medicine at UC San Diego. His research focuses on the complex microbial communities associated with humans, with the environment, and how to scale microbiome analysis to large sample sizes. He been heavily involved in the development of popular computational tools, such as QIIME, Qiita, and PICRUSt, and redeveloped the commonly used phylogenetic algorithm, UniFrac, to support hundreds of thousands of microbiome samples. His publication record in human associated studies includes population scale assessments, examining treatment naive inflammatory bowel disease patients, forensic uses of the microbiome, the identification of novel dietary parameters associated with microbial composition, and integration of multi-omic data. Dr. McDonald received a BS and PhD in Computer Science from the University of Colorado at Boulder.
Twitter, LinkedIn (if desired): @mcdonadt
Bent Petersen
Charla: Examining the diversity of the Amazon rainforest
Position: Associate Professor
Affiliation: Natural History museum of Denmark, Copenhagen University
Bent Petersen is an Associate Professor in bioinformatics at the Natural History museum of Denmark, Copenhagen University. He is also the Deputy Director for the Centre of Excellence for Omics-Driven Computational Biodiscovery (COMBio) at AIMST University, Malaysia. He has a thorough experience in machine learning and has further specialized within the field of genomics and metagenomics where he is working extensively with Next Generation Sequencing data. His research group actively collects samples from all over the world and develops new tools that address the unique challenges of microbial and antique systems biology often by using machine learning.
Currently, he is working extensively on genome and metagenome projects from various branches of the tree of life where they are using supercomputers to analyse the data. In the recent years, drawn by the challenges of plant genome data analysis, Bent has further developed in the field of plant genomics and established the Amazon Rainforest Genome Ontology project (ARGO). A website with video of the project can be seen here: http://www.amazonia.life/.



 FOROS ISIS
FOROS ISIS 















